Our research interests
Regulation of gene expression is fundamental for all eukaryotes. As sessile organisms, plants rely on fast and reliable control mechanisms that regulate gene expression especially under stress conditions. Gene expression can be adjusted and fine-tuned at various levels. We are particularly interested in post-transcriptional mechanisms that ensure survival under stress conditions.
We use Arabidopsis thaliana for most of experiments. Arabidopsis grows across the northern hemiphere and turned out to be a suitable model plant for many research questions. The main advantage of Arabidopsis is that it can easily be genetically manipulated. We use T-DNA and CRISPR/CAS9-induced mutants to study the functions of specific genes during plant development and stress adaptation. Arabidopsis contains more than 25,000 genes, many of which produce more than one RNA isoform.
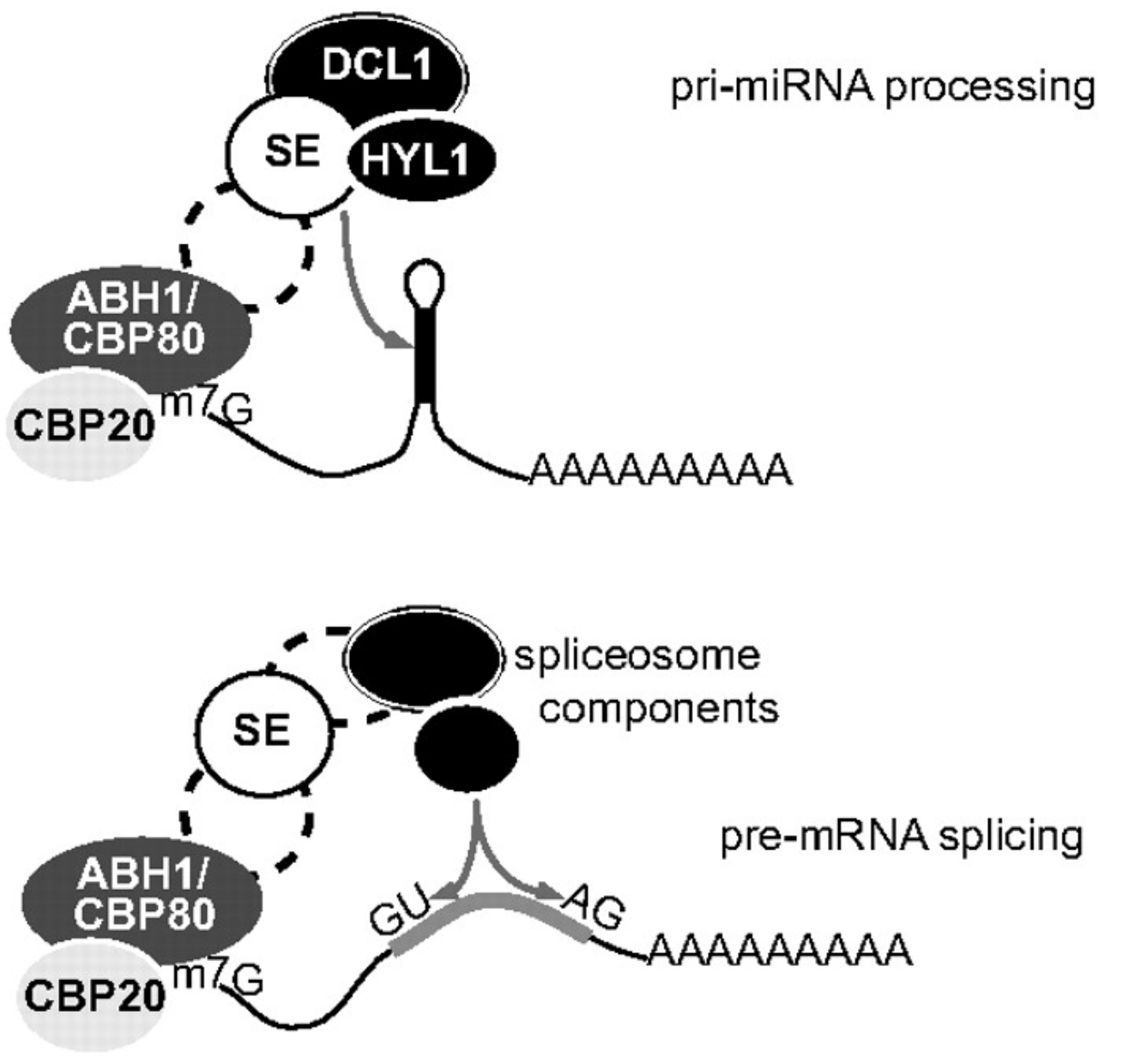
SERRATE regulates RNA processing and transcription
The SERRATE (SE) protein is an essential, conserved eukaryotic RNA processing factor important for plant development. Plant and animal SE are probably best known for its function in the microRNA pathway. But SE also participates in other RNA maturation steps, including constitutive and alternative splicing of mRNAs, RNA 3ʹ-end formation, biogenesis of long non-coding RNAs, RNA transport and RNA stability. The SE protein is composed of different domains, which function in RNA binding and mediate protein-protein interactions. It is still surprising for us that a single protein can fulfill so many different function. We think that specific SE interactors help SE conducting all these specific functions. For instance, SE interacts with HYL and DCL1 to promote microRNA processing. Identifing more of such interactors will eventually help to understand SE function in plants in animals.
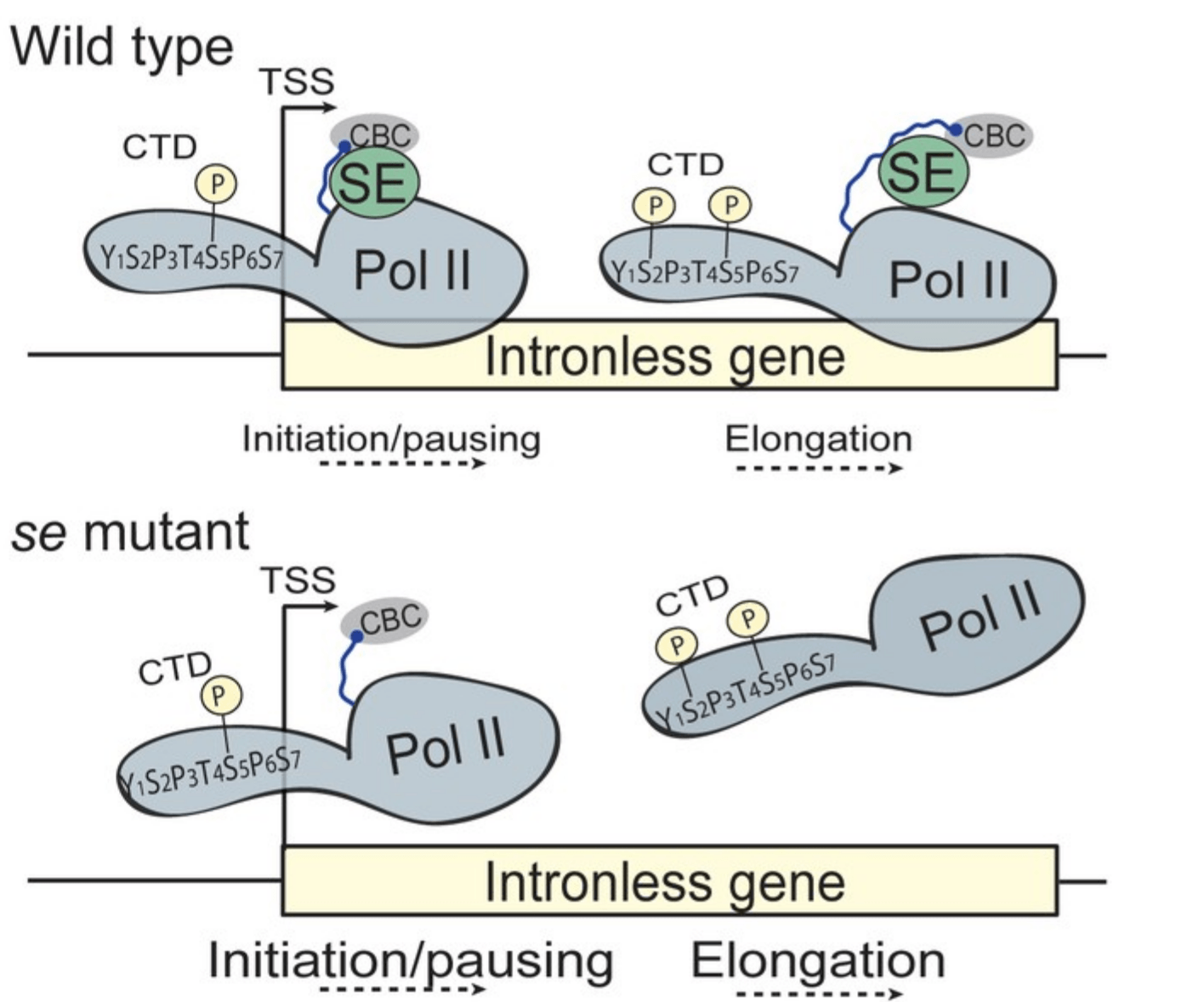
The splicing machinery: The functions of the U1 snRNP in plants
Introns are removed by the spliceosome, a large macromolecular complex composed of five ribonucleoprotein subcomplexes (U snRNPs). The U1 snRNP, which binds to 5ʹ splice sites, plays an essential role in early steps of the splicing reaction. The function of the plant U1 snRNP is not well characterized. We interested in the composition of the plant U1 snRNP and the functions of its subunits for plant development and stress resistance.
We have recently shown that a U1 snRNP accessory component, LUC7, regulates alternative splicing in Arabidopsis. Plants without LUC7 exhibit a dwarf phenotype and react more sensitively when exposed to salt stress or cold. We found hundreds of alternative splicing events in LUC7 mutants. But the most surprising observation was that LUC7 function is particularly important for splicing of terminal introns. LUC7 complexes contain many interesting proteins proteins, such as kinases, which we investigate more deeply.
Recently, we purified the U1 snRNP from plants and analyze their composition by mass spectrometry. Among the proteins that interact with the U1 snRNP, we found several proteins involved in mRNA cleavage and polyadenylation, processes that are crucial for defining the mRNA 3'-end and consequently for mRNA stability and translation efficiency. Our findings reveal that the disruption of core U1 snRNP components in Arabidopsis leads to aberrant premature cleavage and polyadenylation within gene bodies, as well as the selection of alternative polyadenylation sites in 3’ untranslated regions (3'-UTRs). These results highlight the intricate role of U1 snRNP beyond splicing, particularly in the regulation of mRNA 3'-end processing.




Landscape transcriptomics
While most of our studies are conducted under extremely luxurious conditions for plant growth (meaning there are no limitations on water, light, nutrients, and pathogens are mostly absent), life is different for Arabidopsis thaliana in the field. We employ landscape transcriptomic approaches to study the functions of genes in the wild. You can more information here.
